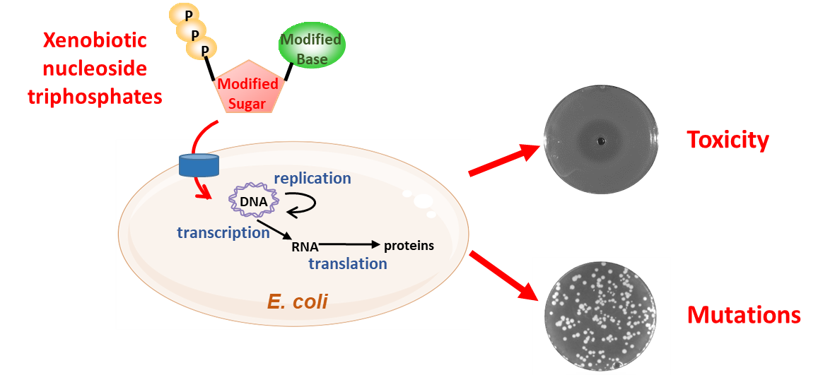
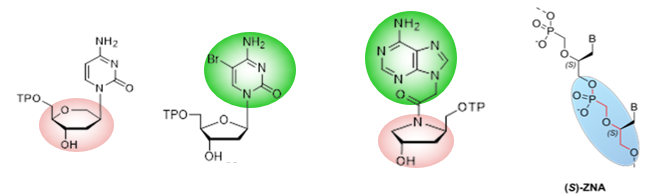
Since the natural genetic code of all life on our planet is recognized only as polymers of DNA and RNA, a robust approach is to develop a truly orthogonal nucleic acid, chemically distinct from DNA and RNA, but which can harbor structural and/or sequence information essential to cell viability and phenotype. Such a Xenobiotic Nucleic Acids (XNA) will be genetically inert, and can only be taken over by artificial enzymes and synthesized from its xenonucleotide (XN) precursors. The latter, not being present in nature, must be chemically synthesized and explicitly supplied to the cell in order to survive. Such “safe” GMOs open new horizons for biotechnological and industrial applications; as non-natural life forms, they also contribute to a better understanding of natural organisms.
XNAs in artificial systems
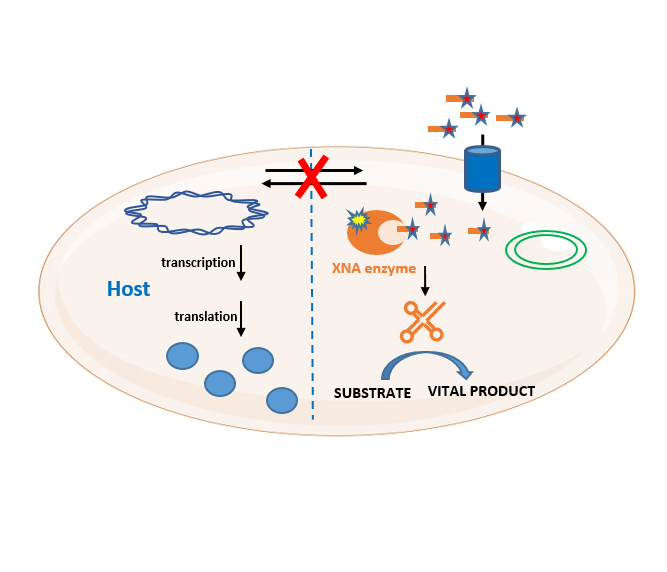
Our objective is to develop an artificial information system based on a new type of nucleic acids that could function in vivo independently of the natural information system. To achieve this goal, four steps have to be elaborated: selection of the backbone chemistry, development of an uptake system in E. coli, evolution of polymerases and selection of vital products.
The Xenome team designs, builds and evolves microorganisms with artificial genetic systems, whose information is carried by XNAs. The proliferation and activity of these organisms are conditioned to the supply of XNAs, placing them under the strict control of the user.
Nucleic acids diversification in Nature
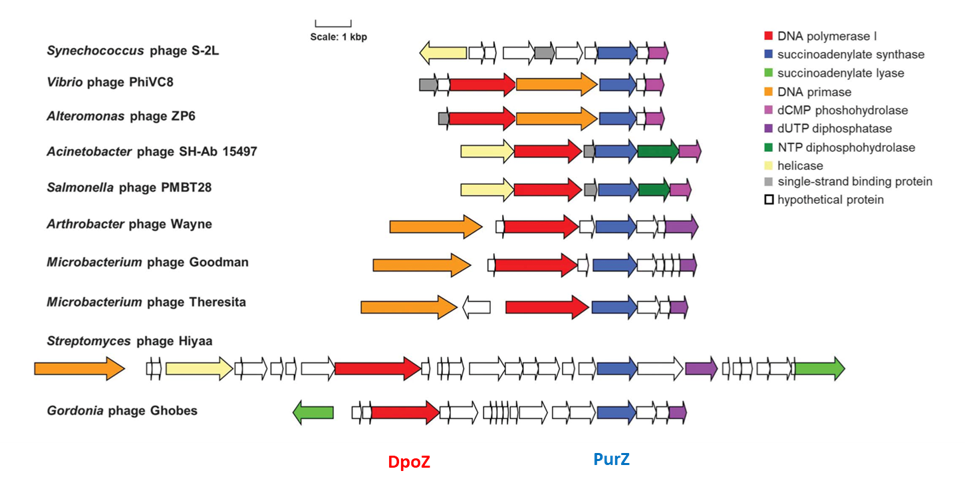
We also study nucleic acid diversification in the natural living world, focusing on bacteriophage genomes that exhibit a high chemical diversity of non-canonical nucleic bases. Some viruses infecting hosts such as proteobacteria, cyanobacteria and actinobacteria have a DNA genome in which adenine is completely replaced by aminoadenine (Z). The elucidation of the biosynthetic pathway of this non-canonical nucleotide and the discovery of bacteriophage DpoZ DNA polymerases dedicated to the incorporation of this nucleotide into DNA pave the way for the propagation of chemically modified genomes in bacteria (Pezo et al., Science 2021).
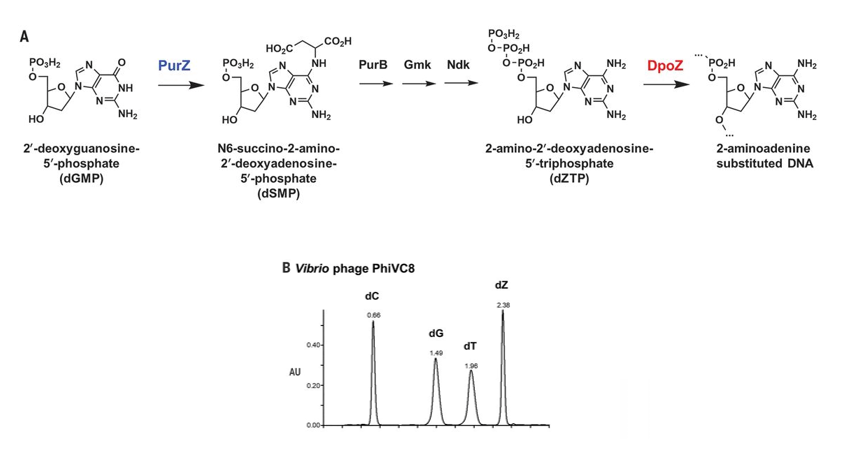
A _ Incorporation pathway of dZTP via reactions catalyzed by the phage-encoded biosynthetic enzyme PurZ and DNA polymerase DpoZ. The steps labeled “PurB,” “Gmk,” and “Ndk” are catalyzed by enzymes of the bacterial host.
B _ HPLC profile of phage DNA digested by nuclease and phosphatase. AU, arbitrary units; dC, deoxycytidine; dG, deoxyguanosine; dT, deoxythymidine; dZ, deoxyaminoadenosine; dA, deoxyadenosine.
Figures extracted from Pezo et al., Science 2021